The Python Hyperspectral Analysis Tool (PyHAT) provides access to data processing, analysis, and machine learning capabilities for spectroscopic applications. It includes a GUI so you can get straight to analyzing data without writing any code. Or, if you are comfortable writing code, PyHAT can be imported just like any other Python package.

Data Format
PyHAT is built on a simple .csv data format that is read into a Pandas data frame for maximum flexibility. Each row of the data frame contains one spectrum and associated metadata and compositional information if available. Each column uses a two-level labeling system which allows the user to do coarse (such as selecting all spectra) or fine (such as selecting the column corresponding to a single wavelength) data selection for simple access to the data.

Hyperspectral Cubes
PyHAT works with hyperspectral cubes by flattening them to match the tabular PyHAT format. Geographic information is stored as metadata so the cube can be reconstructed as needed. PyHAT has built-in support for reading and generating summary parameters from Moon Mineralogy Mapper (M3) and the Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) data cubes. Other data formats can be used if flattened into the PyHAT format, and future work will add more support for common data sets.

Preprocessing
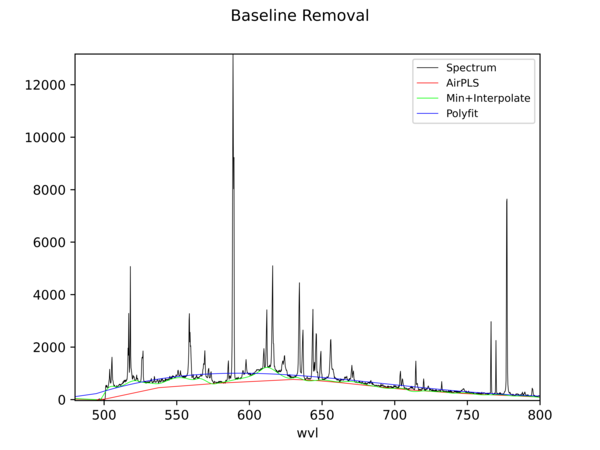
Baseline Removal
PyHAT includes multiple baseline removal algorithms that can be used to remove non-informative signal and leave only the useful features of a spectrum. Built-in plotting functions allow the user to visualize how these algorithms are working and decide which is most suitable.
Dimensionality Reduction
PyHAT includes a variety of dimensionality reduction methods from scikit-learn and other libraries, including:
- Principal Component Analysis (PCA)
- Independent component analysis (ICA)
- t-distributed Stochastic Neighbor Embedding (tSNE)
- Locally Linear Embedding (LLE)
- Non-Negative Matrix Factorization (NNMF)
- Linear Discriminant Analysis (LDA)
- Minimum Noise Fraction (MNF)
- Local Fisher's Discriminant Analysis (LFDA)

Clustering
Both K-Means and Spectral clustering methods are available in PyHAT. Clusters are stored as metadata columns, and can be used in plotting functions to color-code points.

Outlier Identification
PyHAT leverages the Scikit-Learn library for many capabilities., including outlier identification. Two algorithms for outlier identification are implemented: Local Outlier Factor (LOF) and Isolation Forest (IF). Once spectra have been flagged as potential outliers, data manipulation functions allow them to be removed or partitioned into a separate data set.

Regression
A significant focus of PyHAT development has been regression: estimation of a quantitative property based on statistical models that have been trained on spectra of known targets. This is the approach used by the ChemCam and SuperCam teams to derive chemical compositions of Mars rocks using LIBS, but it is broadly applicable to many types of data. PyHAT provides easy access to the following regression algorithms:
- Ordinary Least Squares (OLS)
- Partial Least Squares (PLS)
- Least Absolute Shrinkage and Selection Operator (LASSO)
- Ridge Regression
- Elastic Net
- Bayesian Ridge Regression (BRR)
- Automatic Relevance Determination (ARD)
- Least Angle Regression (LARS)
- Orthogonal Matching Pursuit (OMP)
- Support Vector Regression (SVR)
- Gradient Boosting Regression (GBR)
- Local Regression (algorithm developed by the PyHAT team)
Cross Validation
A key step in the process of training a regression model is tuning its hyperparameters. To avoid overfitting, models must be cross-validated by iteratively withholding some of the training data and predicting it as if it is unknown. This allows the user to identify the parameters that will result in the best balance between training set accuracy and generalizability. PyHAT provides a cross validation module that makes this process simple and utilizes parallel processing to run through the often time-consuming cross validation calculations more efficiently.

Prediction
Once model parameters have been tuned, PyHAT provides the ability to perform predictions on novel data. Multiple different regression methods can be optimized and compared, and the plotting functions include settings to produce a "one-to-one" plot comparing predicted vs actual values to aid in model evaluation and comparison. PyHAT also includes the ability to blend the predictions from multiple submodels, so that the benefits of specialized models can be combined (see related publications for more details).

Introduction to the Python Hyperspectral Analysis Tool (PyHAT)
Post-landing major element quantification using SuperCam laser induced breakdown spectroscopy
Recalibration of the Mars Science Laboratory ChemCam instrument with an expanded geochemical database
Improved accuracy in quantitative laser-induced breakdown spectroscopy using sub-models
The Python Hyperspectral Analysis Tool (PyHAT) provides access to data processing, analysis, and machine learning capabilities for spectroscopic applications. It includes a GUI so you can get straight to analyzing data without writing any code. Or, if you are comfortable writing code, PyHAT can be imported just like any other Python package.

Data Format
PyHAT is built on a simple .csv data format that is read into a Pandas data frame for maximum flexibility. Each row of the data frame contains one spectrum and associated metadata and compositional information if available. Each column uses a two-level labeling system which allows the user to do coarse (such as selecting all spectra) or fine (such as selecting the column corresponding to a single wavelength) data selection for simple access to the data.

Hyperspectral Cubes
PyHAT works with hyperspectral cubes by flattening them to match the tabular PyHAT format. Geographic information is stored as metadata so the cube can be reconstructed as needed. PyHAT has built-in support for reading and generating summary parameters from Moon Mineralogy Mapper (M3) and the Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) data cubes. Other data formats can be used if flattened into the PyHAT format, and future work will add more support for common data sets.

Preprocessing
Baseline Removal
PyHAT includes multiple baseline removal algorithms that can be used to remove non-informative signal and leave only the useful features of a spectrum. Built-in plotting functions allow the user to visualize how these algorithms are working and decide which is most suitable.

Dimensionality Reduction
PyHAT includes a variety of dimensionality reduction methods from scikit-learn and other libraries, including:
- Principal Component Analysis (PCA)
- Independent component analysis (ICA)
- t-distributed Stochastic Neighbor Embedding (tSNE)
- Locally Linear Embedding (LLE)
- Non-Negative Matrix Factorization (NNMF)
- Linear Discriminant Analysis (LDA)
- Minimum Noise Fraction (MNF)
- Local Fisher's Discriminant Analysis (LFDA)

Clustering
Both K-Means and Spectral clustering methods are available in PyHAT. Clusters are stored as metadata columns, and can be used in plotting functions to color-code points.

Outlier Identification
PyHAT leverages the Scikit-Learn library for many capabilities., including outlier identification. Two algorithms for outlier identification are implemented: Local Outlier Factor (LOF) and Isolation Forest (IF). Once spectra have been flagged as potential outliers, data manipulation functions allow them to be removed or partitioned into a separate data set.

Regression
A significant focus of PyHAT development has been regression: estimation of a quantitative property based on statistical models that have been trained on spectra of known targets. This is the approach used by the ChemCam and SuperCam teams to derive chemical compositions of Mars rocks using LIBS, but it is broadly applicable to many types of data. PyHAT provides easy access to the following regression algorithms:
- Ordinary Least Squares (OLS)
- Partial Least Squares (PLS)
- Least Absolute Shrinkage and Selection Operator (LASSO)
- Ridge Regression
- Elastic Net
- Bayesian Ridge Regression (BRR)
- Automatic Relevance Determination (ARD)
- Least Angle Regression (LARS)
- Orthogonal Matching Pursuit (OMP)
- Support Vector Regression (SVR)
- Gradient Boosting Regression (GBR)
- Local Regression (algorithm developed by the PyHAT team)
Cross Validation
A key step in the process of training a regression model is tuning its hyperparameters. To avoid overfitting, models must be cross-validated by iteratively withholding some of the training data and predicting it as if it is unknown. This allows the user to identify the parameters that will result in the best balance between training set accuracy and generalizability. PyHAT provides a cross validation module that makes this process simple and utilizes parallel processing to run through the often time-consuming cross validation calculations more efficiently.

Prediction
Once model parameters have been tuned, PyHAT provides the ability to perform predictions on novel data. Multiple different regression methods can be optimized and compared, and the plotting functions include settings to produce a "one-to-one" plot comparing predicted vs actual values to aid in model evaluation and comparison. PyHAT also includes the ability to blend the predictions from multiple submodels, so that the benefits of specialized models can be combined (see related publications for more details).


